Simplifying Decision Tree Interpretability with Python & Scikit-learn
This post will look at a few different ways of attempting to simplify decision tree representation and, ultimately, interpretability. All code is in Python, with Scikit-learn being used for the decision tree modeling.
When discussing classifiers, decision trees are often thought of as easily interpretable models when compared to numerous more complex classifiers, especially those of the blackbox variety. And this is generally true.
This is especially true of rather comparatively simple models created from simple data. This is much-less true of complex decision trees crafted from large amounts of (high-dimensional) data. Even otherwise straightforward decision trees which are of great depth and/or breadth, consisting of heavy branching, can be difficult to trace.
Concise, textual representations of decision trees can often nicely summarize decision tree models. Additionally, certain textual representations can have further use beyond their summary capabilities. For example, automatically generating functions with the ability to classify future data by passing instances to such functions may be of use in particular scenarios. But let's not get off course -- interpretability is the goal of what we are discussing here.
This post will look at a few different ways of attempting to simplify decision tree representation and, ultimately, interpretability. All code is in Python, with Scikit-learn being used for the decision tree modeling.
Building a Classifier
First off, let's use my favorite dataset to build a simple decision tree in Python using Scikit-learn's decision tree classifier, specifying information gain as the criterion and otherwise using defaults. Since we aren't concerned with classifying unseen instances in this post, we won't bother with splitting our data, and instead just construct a classifier using the dataset in its entirety.
import numpy as np
from sklearn import datasets
from sklearn import tree
# Load iris
iris = datasets.load_iris()
X = iris.data
y = iris.target
# Build decision tree classifier
dt = tree.DecisionTreeClassifier(criterion='entropy')
dt.fit(X, y)
Representing the Model Visually
One of the easiest ways to interpret a decision tree is visually, accomplished with Scikit-learn using these few lines of code:
dotfile = open("dt.dot", 'w')
tree.export_graphviz(dt, out_file=dotfile, feature_names=iris.feature_names)
dotfile.close()
Copying the contents of the created file ('dt.dot' in our example) to a graphviz rendering agent, we get the following representation of our decision tree:
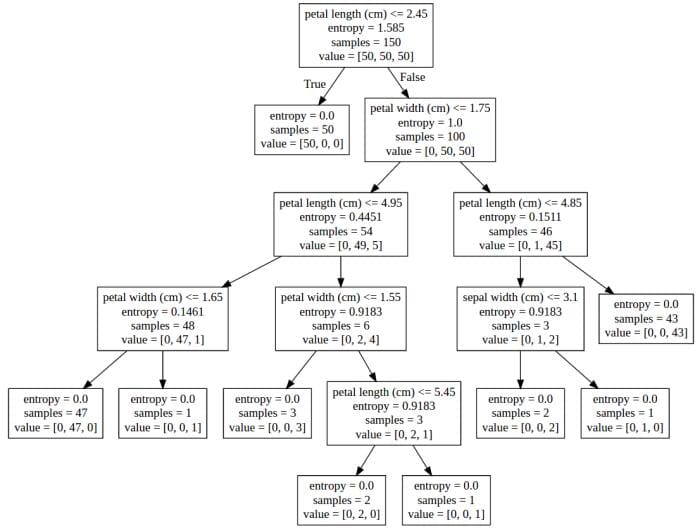
Visual representation of our decision tree using a graphviz rendering agent.
Representing the Model as a Function
As stated at the outset of this post, we will look at a couple of different ways for textually representing decision trees.
The first is representing the decision tree model as a function.
from sklearn.tree import _tree
def tree_to_code(tree, feature_names):
"""
Outputs a decision tree model as a Python function
Parameters:
-----------
tree: decision tree model
The decision tree to represent as a function
feature_names: list
The feature names of the dataset used for building the decision tree
"""
tree_ = tree.tree_
feature_name = [
feature_names[i] if i != _tree.TREE_UNDEFINED else "undefined!"
for i in tree_.feature
]
print("def tree({}):".format(", ".join(feature_names)))
def recurse(node, depth):
indent = " " * depth
if tree_.feature[node] != _tree.TREE_UNDEFINED:
name = feature_name[node]
threshold = tree_.threshold[node]
print("{}if {} <= {}:".format(indent, name, threshold))
recurse(tree_.children_left[node], depth + 1)
print("{}else: # if {} > {}".format(indent, name, threshold))
recurse(tree_.children_right[node], depth + 1)
else:
print("{}return {}".format(indent, tree_.value[node]))
recurse(0, 1)
Let's call this function and see the results:
tree_to_code(dt, list(iris.feature_names))
def tree(sepal length (cm), sepal width (cm), petal length (cm), petal width (cm)):
if petal length (cm) <= 2.45000004768:
return [[ 50. 0. 0.]]
else: # if petal length (cm) > 2.45000004768
if petal width (cm) <= 1.75:
if petal length (cm) <= 4.94999980927:
if petal width (cm) <= 1.65000009537:
return [[ 0. 47. 0.]]
else: # if petal width (cm) > 1.65000009537
return [[ 0. 0. 1.]]
else: # if petal length (cm) > 4.94999980927
if petal width (cm) <= 1.54999995232:
return [[ 0. 0. 3.]]
else: # if petal width (cm) > 1.54999995232
if petal length (cm) <= 5.44999980927:
return [[ 0. 2. 0.]]
else: # if petal length (cm) > 5.44999980927
return [[ 0. 0. 1.]]
else: # if petal width (cm) > 1.75
if petal length (cm) <= 4.85000038147:
if sepal length (cm) <= 5.94999980927:
return [[ 0. 1. 0.]]
else: # if sepal length (cm) > 5.94999980927
return [[ 0. 0. 2.]]
else: # if petal length (cm) > 4.85000038147
return [[ 0. 0. 43.]]
Interesting. Let's see if we can improve interpretability by stripping away some of the "functionality," provided it is not required.
Representing the Model as Pseudocode
Next, a slight reworking of the above code results in the promised goal of this post's title: a set of decision rules for representing a decision tree, in slightly less-Pythony pseudocode.
def tree_to_pseudo(tree, feature_names):
"""
Outputs a decision tree model as if/then pseudocode
Parameters:
-----------
tree: decision tree model
The decision tree to represent as pseudocode
feature_names: list
The feature names of the dataset used for building the decision tree
"""
left = tree.tree_.children_left
right = tree.tree_.children_right
threshold = tree.tree_.threshold
features = [feature_names[i] for i in tree.tree_.feature]
value = tree.tree_.value
def recurse(left, right, threshold, features, node, depth=0):
indent = " " * depth
if (threshold[node] != -2):
print(indent,"if ( " + features[node] + " <= " + str(threshold[node]) + " ) {")
if left[node] != -1:
recurse (left, right, threshold, features, left[node], depth+1)
print(indent,"} else {")
if right[node] != -1:
recurse (left, right, threshold, features, right[node], depth+1)
print(indent,"}")
else:
print(indent,"return " + str(value[node]))
recurse(left, right, threshold, features, 0)
Let's test this function:
tree_to_pseudo(dt, list(iris.feature_names))
if ( petal length (cm) <= 2.45000004768 ) {
return [[ 50. 0. 0.]]
} else {
if ( petal width (cm) <= 1.75 ) {
if ( petal length (cm) <= 4.94999980927 ) {
if ( petal width (cm) <= 1.65000009537 ) {
return [[ 0. 47. 0.]]
} else {
return [[ 0. 0. 1.]]
}
} else {
if ( petal width (cm) <= 1.54999995232 ) {
return [[ 0. 0. 3.]]
} else {
if ( petal length (cm) <= 5.44999980927 ) {
return [[ 0. 2. 0.]]
} else {
return [[ 0. 0. 1.]]
}
}
}
} else {
if ( petal length (cm) <= 4.85000038147 ) {
if ( sepal length (cm) <= 5.94999980927 ) {
return [[ 0. 1. 0.]]
} else {
return [[ 0. 0. 2.]]
}
} else {
return [[ 0. 0. 43.]]
}
}
}
This looks pretty good as well, and — in my computer science-trained mind — the use of well-placed C-style braces makes this a bit more legible then the previous attempt.
These gems have made me want to modify code to get to true decision rules, which I plan on playing with after finishing this post. If I get anywhere of note, I will return here and post my findings.
Matthew Mayo (@mattmayo13) is a Data Scientist and the Editor-in-Chief of KDnuggets, the seminal online Data Science and Machine Learning resource. His interests lie in natural language processing, algorithm design and optimization, unsupervised learning, neural networks, and automated approaches to machine learning. Matthew holds a Master's degree in computer science and a graduate diploma in data mining. He can be reached at editor1 at kdnuggets[dot]com.
